mammos_spindynamics quickstart#
mammos_spindynamics provides an interface for working with atomistic spin-dynamics simulations and data.
dbcontains pre-computed temperature-dependent spontaeous magnetization values for several materialsuppasdprovides a Python interface to UppASD
import os
import shutil
import mammos_dft
import mammos_spindynamics
os.environ.setdefault("OMP_NUM_THREADS", "1"); # Use only 1 thread (needed for Binder)
Querying the database – mammos_spindynamics.db#
Use the following function to get a list of all available materials:
mammos_spindynamics.db.find_materials()
| chemical_formula | space_group_name | space_group_number | cell_length_a | cell_length_b | cell_length_c | cell_angle_alpha | cell_angle_beta | cell_angle_gamma | cell_volume | ICSD_label | OQMD_label | label | source | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Co2Fe2H4 | P6_3/mmc | 194 | 2.645345 Angstrom | 2.645314 Angstrom | 8.539476 Angstrom | 90.0 deg | 90.0 deg | 120.0 deg | 51.751119 Angstrom3 | 0001 | Uppsala | ||
| 1 | Y2Ti4Fe18 | P4/mbm | 127 | 8.186244 Angstrom | 8.186244 Angstrom | 4.892896 Angstrom | 90.0 deg | 90.0 deg | 90.0 deg | 327.8954234 Angstrom3 | 0002 | Uppsala | ||
| 2 | Nd2Fe14B | P42/mnm | 136 | 8.78 Angstrom | 8.78 Angstrom | 12.12 Angstrom | 90.0 deg | 90.0 deg | 90.0 deg | 933.42 Angstrom3 | 0003 | https://doi.org/10.1103/PhysRevB.99.214409 | ||
| 3 | Fe16N2 | ? | 0 | nan Angstrom | nan Angstrom | nan Angstrom | nan deg | nan deg | nan deg | nan Angstrom3 | 0005 | Uppsala | ||
| 4 | Ni80Fe20 | Pm-3m | 221 | 3.55 Angstrom | 3.55 Angstrom | 3.55 Angstrom | 90.0 deg | 90.0 deg | 90.0 deg | 44.738875 Angstrom3 | 0006 | https://doi.org/10.48550/arXiv.1908.08885; spa... | ||
| 5 | Fe3Y | 0 | 5.088172 Angstrom | 5.088172 Angstrom | 24.355398 Angstrom | 90.0 deg | 90.0 deg | 120.0 deg | 546.071496 Angstrom3 | 0007 | Uppsala | |||
| 6 | Fe2.33Ta0.67Y | 0 | 5.227483 Angstrom | 5.227483 Angstrom | 25.022642 Angstrom | 90.0 deg | 90.0 deg | 120.0 deg | 592.173679 Angstrom3 | 0008 | Uppsala |
Use the following function to get an object that contains spontaneous magnetization Ms at temperatures T:
results_spindynamics = mammos_spindynamics.db.get_spontaneous_magnetization("Co2Fe2H4")
The result object provides a function to plot the data for visual inspection:
results_spindynamics.plot()
<Axes: xlabel='Thermodynamic Temperature (K)', ylabel='Spontaneous Magnetization (A / m)'>
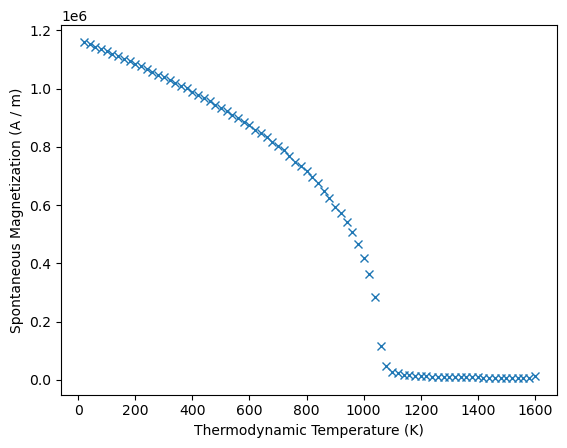
We can access the Ms and T attributes and get mammos_entity.Entity objects:
results_spindynamics.Ms
[1160762.15152728 1152764.540031 1144332.38021977 1136095.4237953
1128134.37709324 1119860.36127139 1111776.43793683 1102918.50364088
1094392.01798628 1085670.06372291 1076778.81897829 1067425.28513189
1057970.97409635 1048538.21055569 1039913.8221449 1029517.07701433
1019718.19960104 1010212.40682623 1000827.01766803 990071.55620798
977847.7971226 967365.03404838 957943.74673504 945368.08824221
933306.33678552 924042.80638082 911156.5370935 898923.20851189
886959.86875088 874610.18656417 859777.5032481 846674.7339648
833341.85234593 817215.36597138 802915.55664802 788770.83767705
767322.55015624 749504.37038855 733715.25355713 716555.57268794
695546.66554008 675931.69422208 650394.96667178 623964.15571257
595267.15471822 574337.71381396 540677.01870906 506480.06838947
465749.37929254 419581.94398809 363556.9147052 283780.87532889
116330.7659029 45920.08375439 28204.38382936 22252.02685696
18336.06448858 16018.35637159 14232.77360447 13649.49689592
12290.85695223 11386.3050879 10781.61435564 10306.54019184
9665.93883184 9395.09234065 8975.25861354 8438.06343775
8343.59372938 8102.70528087 7747.89846694 7593.11190125
7321.35684554 7307.02274373 7047.92251168 7129.16483945
6674.61311621 6599.17726486 6616.13202518 12484.21016472],
unit=A / m)
results_spindynamics.T
[ 20. 40. 60. 80. 100. 120. 140. 160. 180. 200. 220. 240.
260. 280. 300. 320. 340. 360. 380. 400. 420. 440. 460. 480.
500. 520. 540. 560. 580. 600. 620. 640. 660. 680. 700. 720.
740. 760. 780. 800. 820. 840. 860. 880. 900. 920. 940. 960.
980. 1000. 1020. 1040. 1060. 1080. 1100. 1120. 1140. 1160. 1180. 1200.
1220. 1240. 1260. 1280. 1300. 1320. 1340. 1360. 1380. 1400. 1420. 1440.
1460. 1480. 1500. 1520. 1540. 1560. 1580. 1600.],
unit=K)
To work with the data we can also get it as pandas.DataFrame. The dataframe only contains the values (not units).
results_spindynamics.dataframe
| T | Ms | |
|---|---|---|
| 0 | 20.0 | 1.160762e+06 |
| 1 | 40.0 | 1.152765e+06 |
| 2 | 60.0 | 1.144332e+06 |
| 3 | 80.0 | 1.136095e+06 |
| 4 | 100.0 | 1.128134e+06 |
| ... | ... | ... |
| 75 | 1520.0 | 7.129165e+03 |
| 76 | 1540.0 | 6.674613e+03 |
| 77 | 1560.0 | 6.599177e+03 |
| 78 | 1580.0 | 6.616132e+03 |
| 79 | 1600.0 | 1.248421e+04 |
80 rows × 2 columns
Running spindynamics simulations – mammos_spindynamics.uppasd#
The uppasd subpackage provides an Python interface to UppASD (the Uppsala Atomistic Spin Dynamics Software). UppASD has to be installed separately and the executable must be in your PATH. The easiest way is to install UppASD from conda-forge using pixi or conda.
In this notebook we will show how to run a single simulation. We assume that we have all required input files. The uppasd notebook will explain both creating inputs as well as analyzing the resulting data in more detail.
We will run a simulation for Co2Fe2H4. We can get three of the required input files for this material from mammos_dft.db:
exchangecontaining the exchange coupling constants \(J_{ij}\)posfilecontaining the position of the atoms in the unit cellmomfilecontaining the magnetic moments of the atoms in the unit cell
We will first copy the three files to the current directory. This most closely resembles the scenario where you may have your own files. We will also rename exchange to jfile (for no good reason, just to show that we can) and use that name in inpsd.dat further down.
In practice, you should avoid this manual copy when using files from the database in mammos_dft and instead pass the paths to the three files as additional keyword arguments when creating the simulation object, as explain in the uppasd notebook.
To access the data files coming with mammos_dft.db we use the get_uppasd_properties function:
uppasd_inputs = mammos_dft.db.get_uppasd_properties("Co2Fe2H4")
The returned object gives access to the available files, e.g.:
uppasd_inputs.posfile
PosixPath('/home/mlang/repos/mammos/mammos-devtools/packages/mammos-dft/src/mammos_dft/data/0001/posfile')
# copy files to simulate that we have our own files
shutil.copy(uppasd_inputs.posfile, ".")
shutil.copy(uppasd_inputs.momfile, ".")
shutil.copy(uppasd_inputs.exchange, "./jfile");
In addition we have to create the main input file inpsd.dat. For detailed documentation about this file and UppASD in general refer to the UppASD documentation.
In this notebook we create the file using the writefile magic, as a user you may already have your own input file, which you can use.
Warning
In order to keep the runtime of this notebook short we use unreasonably small values for ncell, ip_mcnstep and mcnstep.
For production runs you will have to use higher values, suitable starting points could be ncell 32 32 32, ip_mcnstep 25000, mcnstep 50000
%%writefile inpsd.dat
simid Co2Fe2H4
cell 1.000000000000000 -0.500000000182990 0.000000000000000
0.000000000000000 0.866011964524435 0.000000000000000
0.000000000000000 0.000000000000000 3.228114436804486
ncell 12 12 12
bc P P P
sym 0
posfile ./posfile
posfiletype D
initmag 3
momfile ./momfile
maptype 2
exchange ./jfile
ip_mode M
ip_temp 20
ip_mcnstep 200
mode M
temp 20
mcnstep 200
plotenergy 1
do_proj_avrg Y
do_cumu Y
alat 2.6489381562e-10
Writing inpsd.dat
We pass the input file paths together with paths to the other three files to the Simulation class:
sim = mammos_spindynamics.uppasd.Simulation(inpsd="inpsd.dat")
We can now use this object to run a simulation. We need to pass a path to a directory where the output will be written to. Optionally, we can also pass a human-readable description, which will be saved as additional metadata.
result = sim.run(out="Co2Fe2H4-short", description="Short test for Co2Fe2H4")
Running UppASD in Co2Fe2H4-short/0-run ... simulation finished, took 0:00:07
We get an object back, which allows us to inspect simulation output:
result.T
result.Ms
result.Cv
We can also access some simulation metadata, e.g. the description and the runtime:
result.info()
| name | description | time_elapsed | |
|---|---|---|---|
| 0 | 0-run | Short test for Co2Fe2H4 | 0:00:07 |
